kmerHash
Project apply kmer and minHash method for genome comparions.
# Create a conda env with python=3.7 and install packages.
conda create -n sourmash python=3.7
conda activate sourmash
conda install -c bioconda sourmash
conda install -c bioconda ncbi-genome-download
conda install -c bioconda khmer
# Downlaod genome/fasta files from ncbi
ncbi-genome-download --format fasta --genus "Bacteroides fragilis" bacteria -p 8
Since this will create individaul folder per genome, we can pull them together into a single folder.
# make a symbolic link to the genbank file
mkdir data
## Create a symbolic link of genome files to the data folder
ln -s /Users/ravinpoudel/Documents/sourmash/refseq/bacteria/**/*.fna.gz data/
Now we have the input files/ genomes. We will apply kmer and minHash methods. Create MinHash sketches(signature) at k-mer sizes of 31
time sourmash compute --scaled 1000 -k 31 -p 16 *.fna.gz --outdir Bfrag_many_sig
Let say one of the signature represents a signature assocaited with reference genome. Then if we want to find the closet match of query sigature to the reference then,
reference signature = GCF_003471465.1_ASM347146v1_genomic.fna.gz.sig
mv Bfrag_many_sig/GCF_003471465.1_ASM347146v1_genomic.fna.gz.sig .
Now compare all the signature with the reference genome.
First, Let's turn all the singatures into an easily-searchable database with sourmash index –
sourmash index -k 31 bfragdb Bfrag_many_sig/*.sig
## output == bfragdb.sbt.json
Now we can search against reference.
sourmash search GCF_003471465.1_ASM347146v1_genomic.fna.gz.sig bfragdb.sbt.json -n 20
############## OUTPUT ######################
16 matches:
similarity match
---------- -----
59.9% GCF_000710375.2_ASM71037v3_genomic.fna.gz
59.2% GCF_902364655.1_MGYG-HGUT-00236_genomic.fna.gz
55.1% GCF_000710365.2_ASM71036v3_genomic.fna.gz
53.3% GCF_000297695.1_Bact_frag_HMW_610_V1_genomic.fna.gz
52.8% GCF_003439065.1_ASM343906v1_genomic.fna.gz
48.5% GCF_000724665.2_ASM72466v3_genomic.fna.gz
46.9% GCF_000724815.2_ASM72481v2_genomic.fna.gz
46.8% GCF_000724805.2_ASM72480v2_genomic.fna.gz
45.8% GCF_001695355.1_ASM169535v1_genomic.fna.gz
44.8% GCF_001693695.1_ASM169369v1_genomic.fna.gz
43.7% GCF_002849695.1_ASM284969v1_genomic.fna.gz
43.2% GCF_000157015.1_ASM15701v1_genomic.fna.gz
42.1% GCF_003463545.1_ASM346354v1_genomic.fna.gz
41.7% GCF_000297755.1_Bact_frag_HMW_616_V1_genomic.fna.gz
41.5% GCF_003465265.1_ASM346526v1_genomic.fna.gz
41.4% GCF_003464635.1_ASM346463v1_genomic.fna.gz
Just curious, how well will the match when search within the same genome.
sourmash search GCF_003471465.1_ASM347146v1_genomic.fna.gz.sig GCF_003471465.1_ASM347146v1_genomic.fna.gz.sig
##########OUTPUT ##########
1 matches:
similarity match
---------- -----
100.0% GCF_003471465.1_ASM347146v1_genomic.fna.gz
Compare many signatures and build a tree– application towards comparative genomics.
# Compare all the things, and create comparision plots:
sourmash compare -p 16 Bfrag_many_sig/*.sig -o bfrag_compare
sourmash plot --pdf --labels bfrag_compare
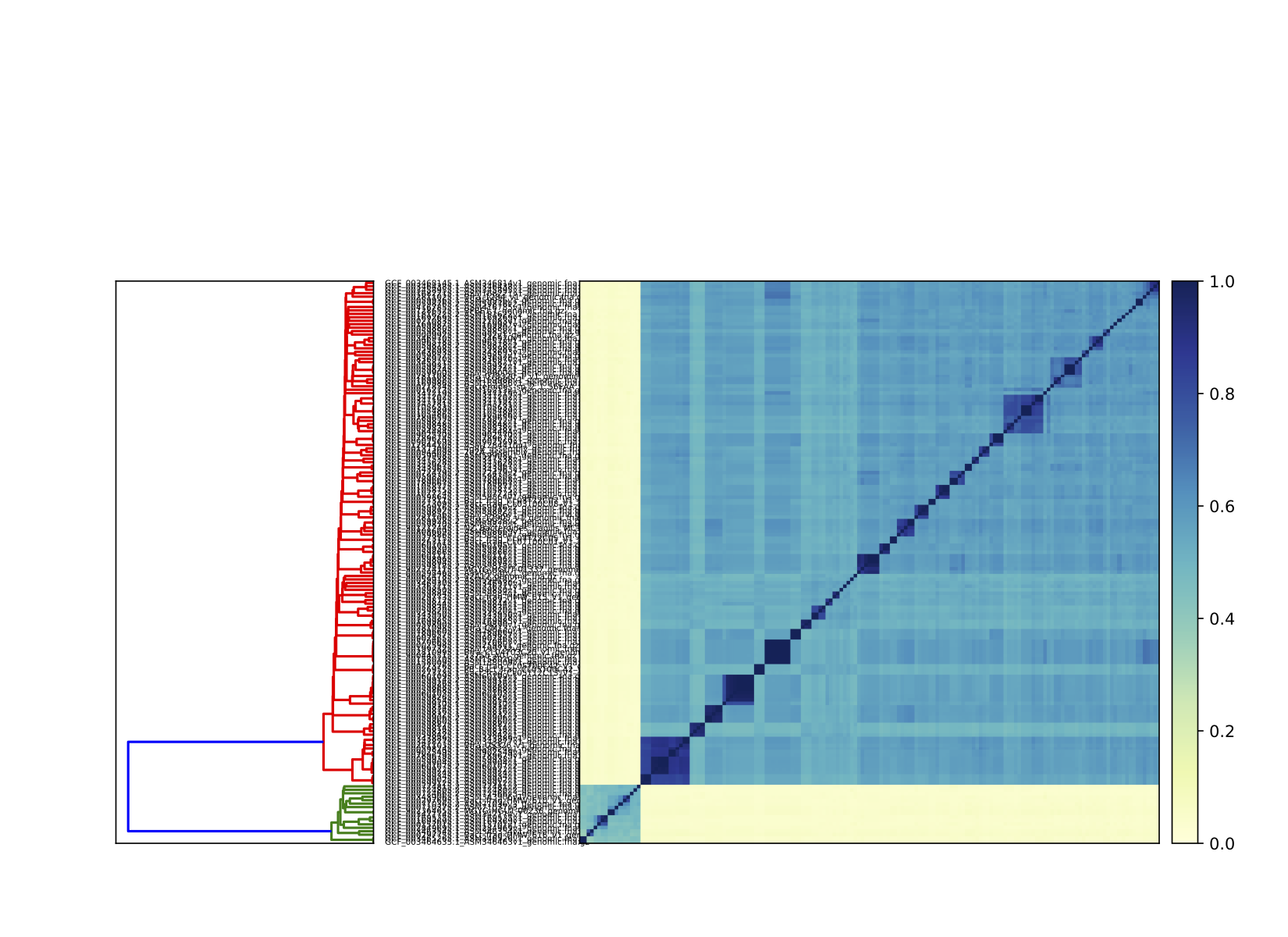
The maximum similarity in the matrix is 1, meaning the two signatures were from the same species at the k-mer size of 31. Some of signature have 0% similarity with other signatures, suggesting the presence of outlier group.
########## Download a database containing all of the GenBank microbial genomes:
curl -L -o genbank-k31.lca.json.gz https://osf.io/4f8n3/download
Next, run the ‘gather' command to see what's in your ecoli genome –
# comparing your fasta/genome across all the GenBank microbial genomes
sourmash gather -k 31 GCF_003471465.1_ASM347146v1_genomic.fna.gz.sig genbank-k31.lca.json.gz
##### output #######
electing specified query k=31
loaded query: GCF_003471465.1_ASM347146v1_ge... (k=31, DNA)
loaded 1 databases.
overlap p_query p_match
--------- ------- -------
4.6 Mbp 76.1% 74.3% JMZY02000001.1 Bacteroides fragilis s...
4.5 Mbp 7.3% 7.4% JMZX02000001.1 Bacteroides fragilis s...
3.9 Mbp 2.5% 2.4% EQ973245.1 Bacteroides fragilis 3_1_1...
4.0 Mbp 1.8% 1.9% JMZZ02000001.1 Bacteroides fragilis s...
4.0 Mbp 1.3% 1.4% JPHS01000001.1 Bacteroides fragilis s...
4.3 Mbp 1.0% 1.0% JH815482.1 Bacteroides fragilis HMW 6...
found less than 50.0 kbp in common. => exiting
found 6 matches total;
the recovered matches hit 90.0% of the query
In the case of metagenomics, you will have multiple metageomes that you want compare across GenBank microbial genomes:
For instanace, lets download metagenome / contig. Lets download a contigs file from HMP2 database(https://ibdmdb.org/tunnel/public/HMP2/WGS/1818/products) - eg- CSM79HLA_TR_contigs.fna
# crate a signatiure file for CSM79HLA_TR_contigs.fna
sourmash compute --scaled 1000 -k 31 -p 16 CSM79HLA_TR_contigs.fna -o CSM79HLA_TR_contigs.sig
# Now compare across GenBank microbial genomes
sourmash gather -k 31 CSM79HLA_TR_contigs.sig genbank-k31.lca.json.gz
## OUTPUT #############
selecting specified query k=31
loaded query: CSM79HLA_TR_contigs.fna... (k=31, DNA)
loaded 1 databases.
overlap p_query p_match
--------- ------- -------
4.4 Mbp 6.8% 73.6% MKXP01000012.1 Bacteroides vulgatus i...
4.1 Mbp 6.3% 73.8% JH724079.1 Bacteroides caccae CL03T12...
4.1 Mbp 6.2% 87.0% CZAD01000001.1 Bacteroides uniformis ...
3.8 Mbp 5.7% 46.8% JH724226.1 Bacteroides ovatus CL02T12...
3.3 Mbp 5.0% 72.3% DS264552.1 Parabacteroides merdae ATC...
2.4 Mbp 3.7% 68.0% KI271434.1 Oscillibacter sp. KLE 1728...
2.3 Mbp 3.6% 61.8% CP003274.1 Alistipes finegoldii DSM 1...
.
.
.
found less than 50.0 kbp in common. => exiting
found 90 matches total;
the recovered matches hit 81.1% of the query
### Export as csv file
sourmash gather -k 31 CSM79HLA_TR_contigs.sig genbank-k31.lca.json.gz -o CSM79HLA_TR_out.csv
Using sourmash LCA to do taxonomic classification. Sourmash LCA is using k-mers to do taxonomic classification, using the "lowest common ancestor" approach.
sourmash lca classify --db genbank-k31.lca.json.gz \
--query GCF_003471465.1_ASM347146v1_genomic.fna.gz.sig
# OUTPUT #########
loaded 1 LCA databases. ksize=31, scaled=10000
finding query signatures...
outputting classifications to -
ID,status,superkingdom,phylum,class,order,family,genus,species,strain
GCF_003471465.1_ASM347146v1_genomic.fna.gz,found,Bacteria,Bacteroidetes,Bacteroidia,Bacteroidales,Bacteroidaceae,Bacteroides,Bacteroides fragilis,
classified 1 signatures total
######
sourmash lca summarize --db genbank-k31.lca.json.gz \
--query GCF_003471465.1_ASM347146v1_genomic.fna.gz.sig
### OUTPUT ########
loaded 1 LCA databases. ksize=31, scaled=10000
finding query signatures...
loaded 1 signatures from 1 files total.
87.7% 529 Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Bacteroidaceae;Bacteroides
87.7% 529 Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Bacteroidaceae
90.7% 547 Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales
90.7% 547 Bacteria;Bacteroidetes;Bacteroidia
90.7% 547 Bacteria;Bacteroidetes
92.4% 557 Bacteria
8.0% 48 Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Bacteroidaceae;Bacteroides;Bacteroides fragilis